BioGPS opened 2016 with a publication in Nucleic Acids Research, right after the New Year holiday. Throughout the year, new designs for the site were being created, reviewed, adjusted, reviewed, adjusted, and more review/adjustments in anticipation of a site redesign for 2017. A Plugin registration Blitz was held in March and April; followed by a Plugin Review Blitz in May. The BioGPS spotlight series was also restarted, with spotlights on BGEE, Intermine, and other Intermine-related plugins.
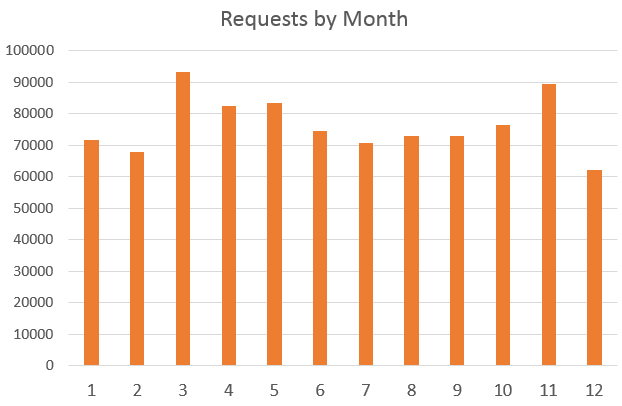
There were ~910,000 requests made to BioGPS in 2016. Requests to BioGPS peaked in March and at the lowest in December.
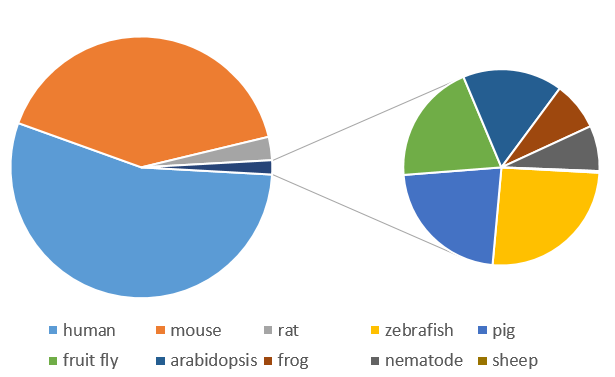
Most searches involved human and mouse genes by far, though thousands of requests were also made for other model organisms.
Discarding the default search genes, CDK2, the top 10 most frequently requested genes were:
- GAPDH
- TP53
- Trpm4
- EGFR
- CD274
- KLF6
- TNF
- ACTB
- MYC
- AKT1
Given their use as controls, the appearance of GAPDH and ACTB in the top 10 list, wasn’t too surprising. Out of the top 10 genes searched in BioGPS, seven had corresponding Gene Wiki articles that were not stubs. The three genes with incomplete Gene Wiki articles were ACTB, Trpm4, and KLF6. If you feel like sharing what you know about these three genes, consider editing their corresponding Wikipedia articles.
BioGPS users may not have been making as many requests in 2016 as they made in 2015, but in 2016, they still published about 72 articles citing the 2009 paper on BioGPS.

Trackbacks/Pingbacks