This is part 2 of a blog post describing Wikidata SPARQL query logs. Part 1 is here. In this part, we’ll look at the most used items and specifically look at diseases, drugs, genes and proteins. And then look at co-occurrence of properties within queries..
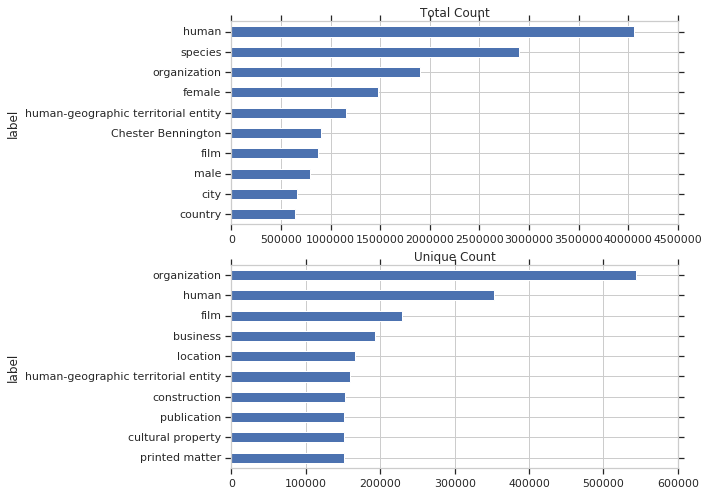
Below is a plot of the most used items by total query count and by unique query count
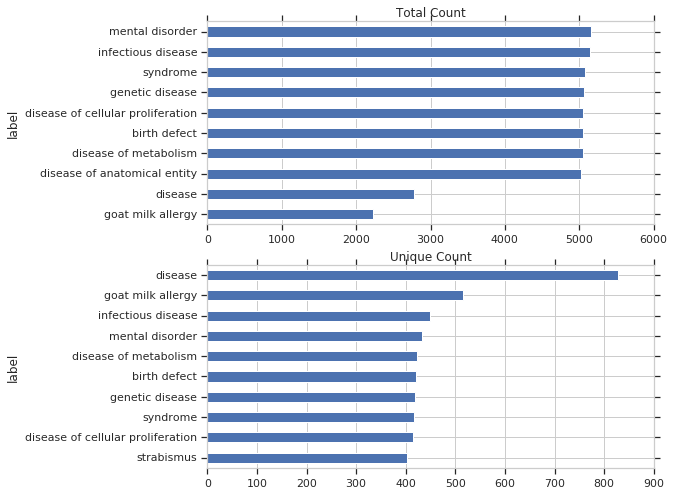
Next, I’ll look specifically at diseases. These are the most used disease items by total query count and by unique query count:
I then retrieved all diseases, drugs, genes and protein from Wikidata and summed the counts for each type. The table below shows some summary information for each. The columns unique and total display the number of unique and total queries containing any item of that type, respectively. The columns organic and robotic contain the number of unique queries classified as organic or robotic, and num_items contains the total number of each type in Wikidata.
| unique | total | organic | robotic | num_items | |
|---|---|---|---|---|---|
| disease | 82388 | 181241 | 1742 | 80465 | 11240 |
| gene | 11795 | 30899 | 163 | 11618 | 757218 |
| protein | 950 | 3798 | 37 | 913 | 533161 |
| drug | 42692 | 209740 | 544 | 42136 | 145961 |
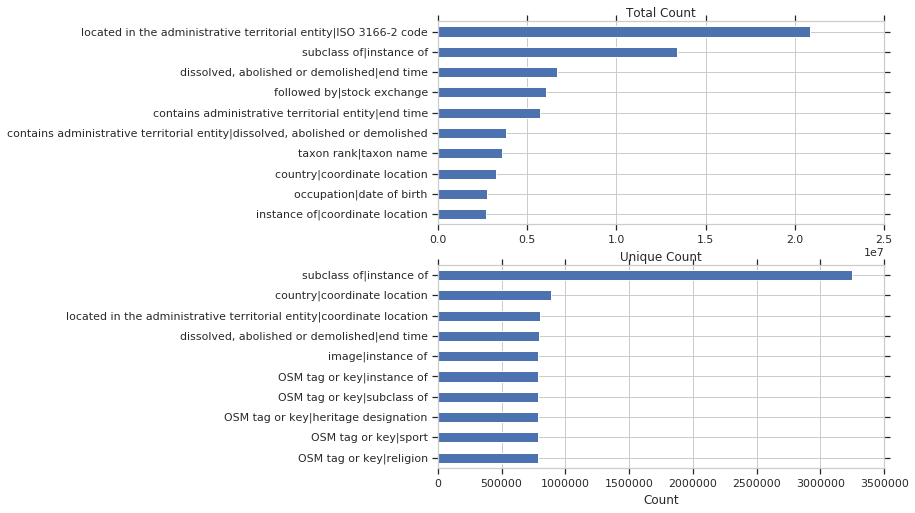
Property Co-occurrence
If two properties are used together within the same query, it may indicate something special.
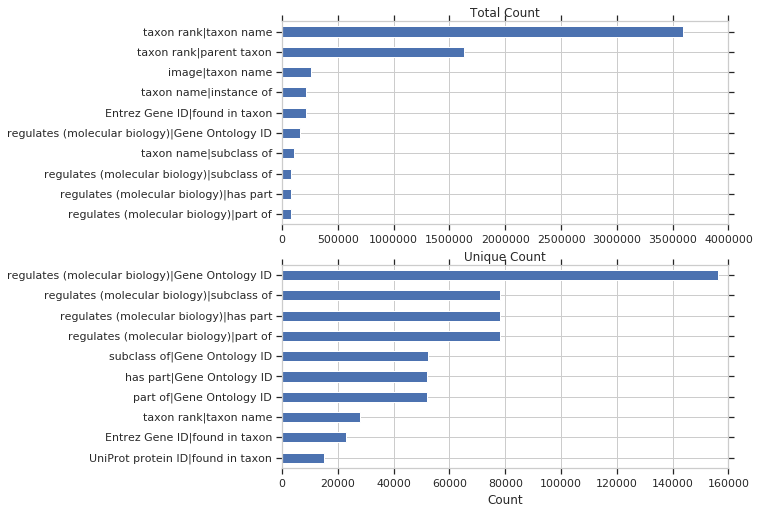
First we’ll look at all properties. Below is a plot of the of top pairs of properties used together in a query, by total and unique count
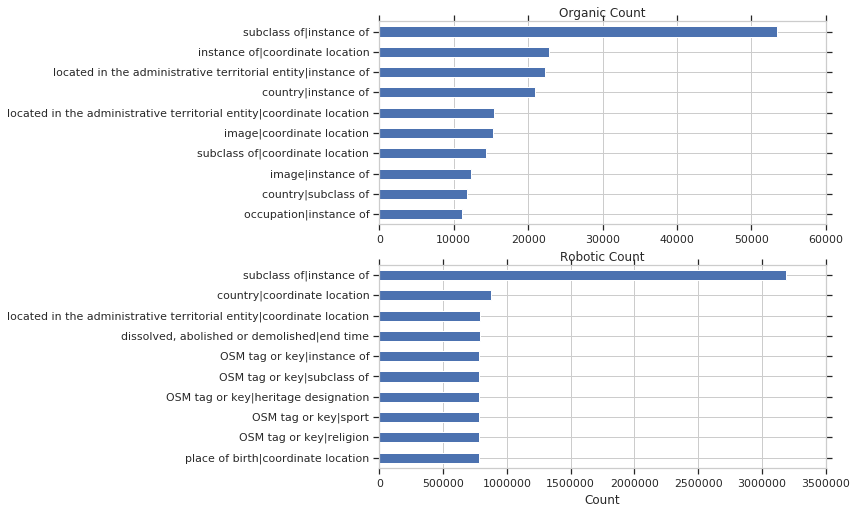
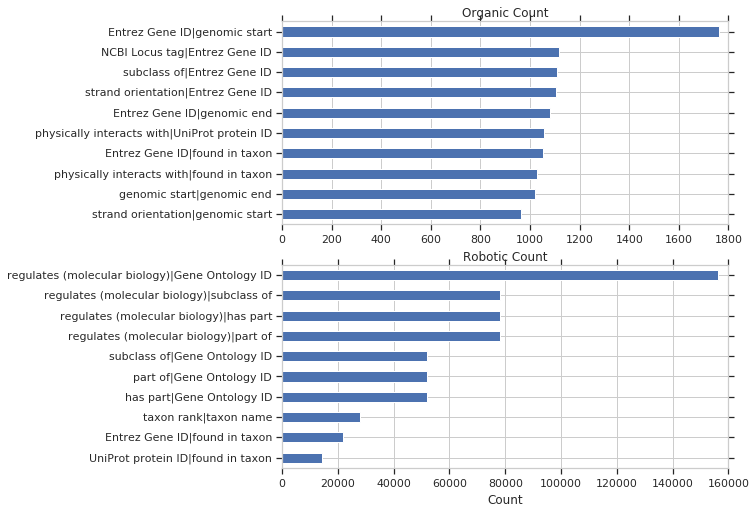
and by organic vs robotic count:
Next, looking specifically at biomedical-related properties:
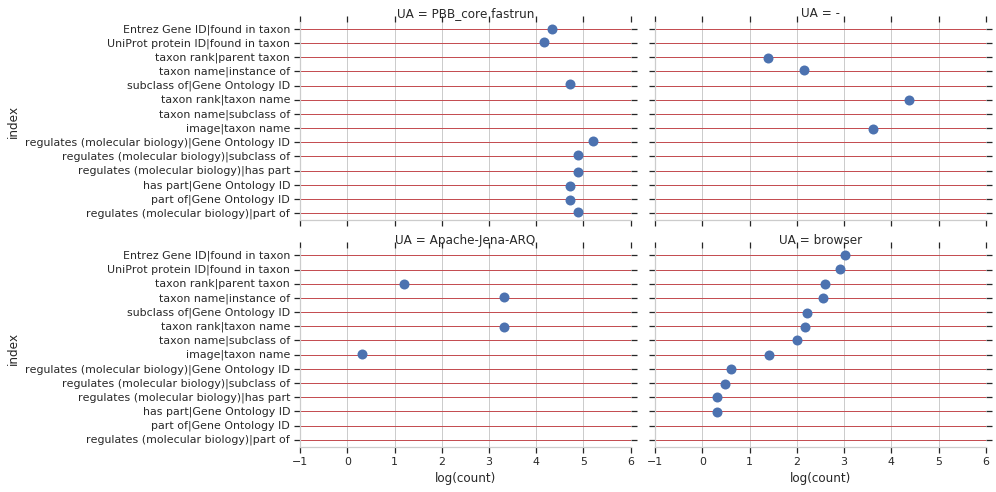
And lastly, I wanted to see the most common useragents for each of these pairs of properties.
These plots together indicates that there were thousands of unique, organic, integrative queries involving biomedical items of different classes performed during this time.