 Bioinformatics web developers often face one common problem — adding a search box that allows users to type a gene, which then directs them to a gene-specific resource page. To implement this feature, developers need to maintain a gene annotation database of their own, and then write the interface to link the user input to the backend query. Moreover, they also need to keep their gene annotation database up to date and stable.
Bioinformatics web developers often face one common problem — adding a search box that allows users to type a gene, which then directs them to a gene-specific resource page. To implement this feature, developers need to maintain a gene annotation database of their own, and then write the interface to link the user input to the backend query. Moreover, they also need to keep their gene annotation database up to date and stable.
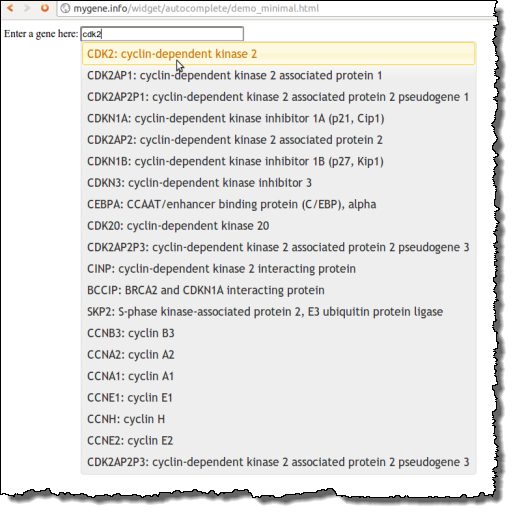
Recently, we developed a web widget to allow developers to enable autocomplete in any input field. This widget is based on Mygene.Info, the same web service engine we built to handle gene queries for BioGPS, so developers don’t even have to deal with that hassle. You can check out the live demo and the instructions here, or the screenshot below:
An example of a minimal HTML page with gene query autocomplete enabled looks just as simple as this:
<html><body>
<label for="gene_query">Enter a gene here: </label>
<input style="width:250px">
<script src="http://mygene.info/widget/autocomplete/js/mygene_query_min.js" type="text/javascript"></script>
</body></html>
For those who are interested in more details of how this is implemented, you can check out the source code here.
For those who haven’t checked out Mygene.Info before, we designed it to provide fast and simple web services for querying and retrieving gene annotations. It easily handles all the gene query traffic from BioGPS, and we’re confident that it can handle whatever traffic you throw at it. We hope these web services can reduce or even eliminate the overheads of many groups each maintaining their own gene annotation databases, all of which essentially come from the same authoritative resources (e.g. NCBI, Ensembl etc).
Give it a spin and let us know what you think!

Just wonder, why there is differnece in expression values in BioGPS and GEO profile website? Also, can I use BioGPS to search for my gene of interest within a specific dataset i.e. GNF1h?