Tools and Infrastruture
Much of our work in building tools and infrastructure focuses on how we as the biomedical scientific community manage data and knowledge. By better organizing information, we hope to improve the efficiency of research and promote data reuse. Several representative resources are described below.

The BioThings suite of application programming interfaces (APIs) focuses on data integration for key biomedical entities, including genes (mygene.info), variants (myvariant.info), and chemicals (mychem.info), which are collectively accessed millions of times per month. We also provide a software development kit (SDK) for easily creating custom APIs, including those served at biothings.transltr.io.
BioThings Explorer (BTE) leverages the semantically-precise annotations of APIs in the SmartAPI registry to create a federated knowledge graph. BTE can autonomously execute multi-hop, heterogeneous queries to explore complex relationships between genes, drugs, diseases, and more. BTE is developed as part of the NCATS Biomedical Data Translator consortium.

The NIAID Data Ecosystem Discovery Portal enables users to find datasets from a wide range of repositories and offers a convenient one-stop-shop for discovery of data on infectious and immune-mediated diseases (IIDs). The Discovery Portal regularly collects metadata from all included repositories and indexes it in a searchable catalogue for anyone to explore.
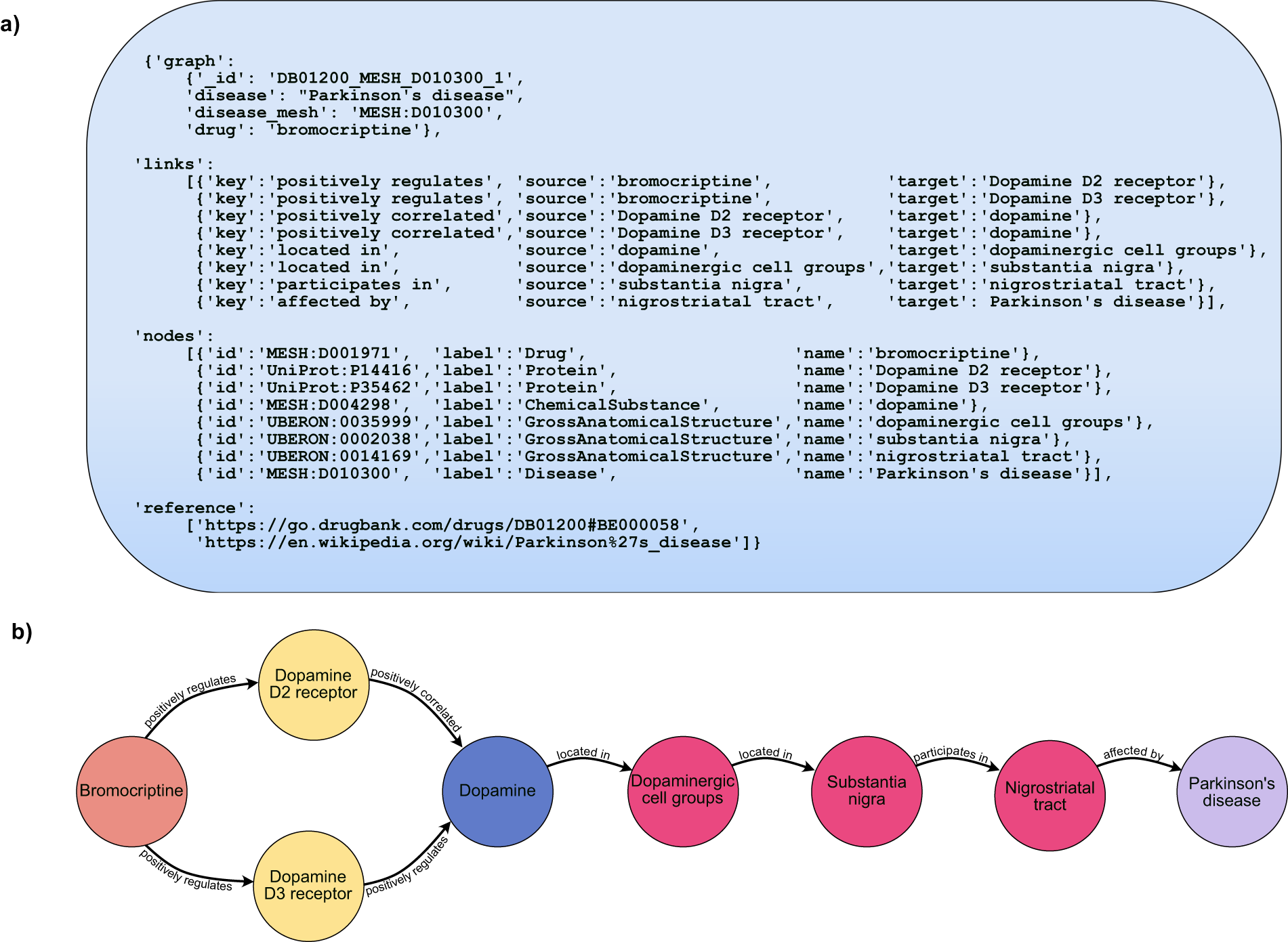
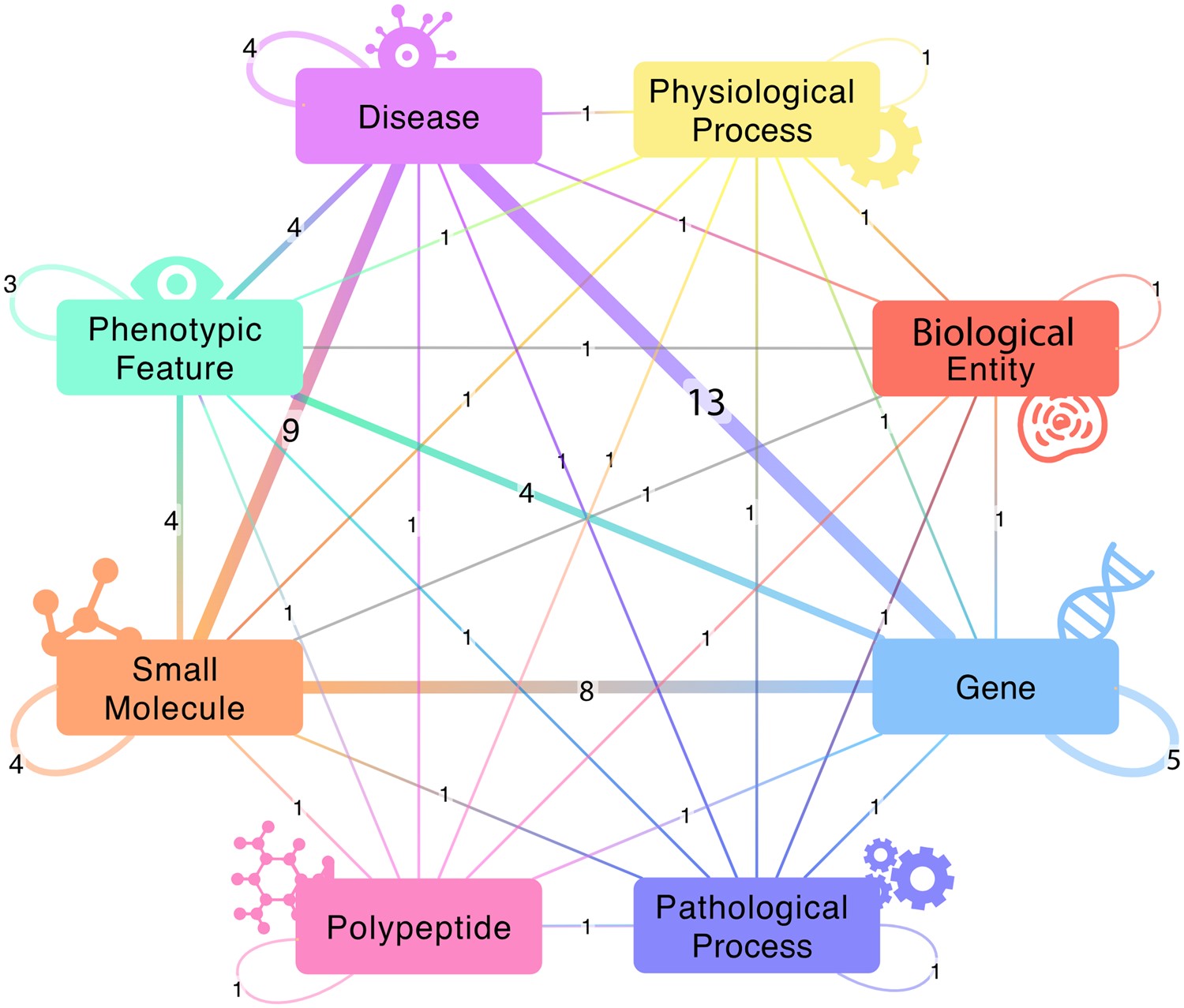
Although there are many databases of drug indications, DrugMechDB is the only database that focuses on drug mechanisms of action. These mechanisms of action are expressed as paths through a biomedical knowledge graph. This resource has been used to train and validate algorithms for drug repurposing.
