Last month, we couldn’t resist highlighting the plugin for an awesome resource which provided an excellent visualization of gene expression/distribution based on multiple data sources. As it turns out, TISSUES is just one of many inspired tools coming out of the Jensen Lab. While we were busy marveling at TISSUES and looking up various genes just to see how TISSUES would display them in our BioGPS layout, Dr. Lars Juhl Jensen quietly added a plugin to one of his other tools–COMPARTMENTS. Without further ado, we introduce COMPARTMENTS! Dr. Lars Juhl Jensen, the PI behind this awesome resource, kindly answered our inquiries.
- In one tweet or less, introduce us to your tool.
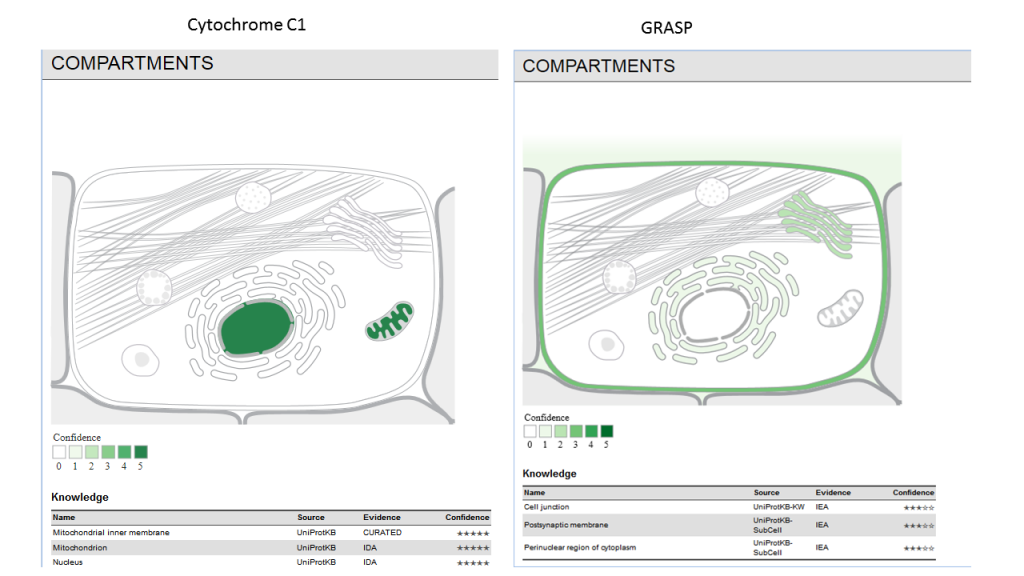
Subcellular localization derived from curated knowledge, high-throughput screens, automatic text mining, and sequence-based predictions.
- Why is your tool unique and special?
Other resources have previously been developed that did much the same as COMPARTMENTS; however, these generally ceased to be updated. In contrast, we designed our infrastructure with a strong focus on automation and synergy with other resources to make it maintainable..
- Who is your target audience?
COMPARTMENTS currently caters to two audiences. The web interface is designed to make it easy for users who simply want to look up individual proteins to get a comprehensive overview of their subcellular localization. The download files, are intended primarily for bioinformaticians, who want to use the full data collection for large-scale studies.
- Why did you create your tool?
Having an overview of where proteins are localized in cells is useful both in my groups work on protein networks and in our collaborations with neighboring proteins groups. Since no existing resource did exactly what we needed, we decided to make our own.
- What is your greatest success story so far?
The sudden growth in users when COMPARTMENTS was included in GeneCards compared to when the article came out was quite an eye-opener.
- What improvements are coming in the future?
We will soon roll out a scoring scheme that calculates the combined confidence score of each localization given all the available evidence. We also consider adding functionality for gene set enrichment analysis and upgrades to the prediction methods used.
- Who is the team behind your knowledge base?
The tool was developed in my group at the Novo Nordisk Foundation Center for Protein Research, University of Copenhagen. We collaborated with the groups of Reinhard Schneider at University of Luxembourg and Seán O’Donoghue at CSIRO/Garvan Institute of Medical Research.
Thanks to Dr. Jensen, for guiding us through their extremely useful and FREE tool. Be sure to check out their plugin in the plugin library.