It’s been awhile since we posted something for our Spotlight series; however, we couldn’t pass up profiling an awesome resource like this one. Given the number questions posted to the BioGPS google groups about gene expression and distribution, TISSUES is resource that is sure to be a hit in the BioGPS community. Dr. Lars Juhl Jensen, the PI behind this awesome resource, kindly answered our inquiries.
- In one tweet or less, introduce us to your tool.
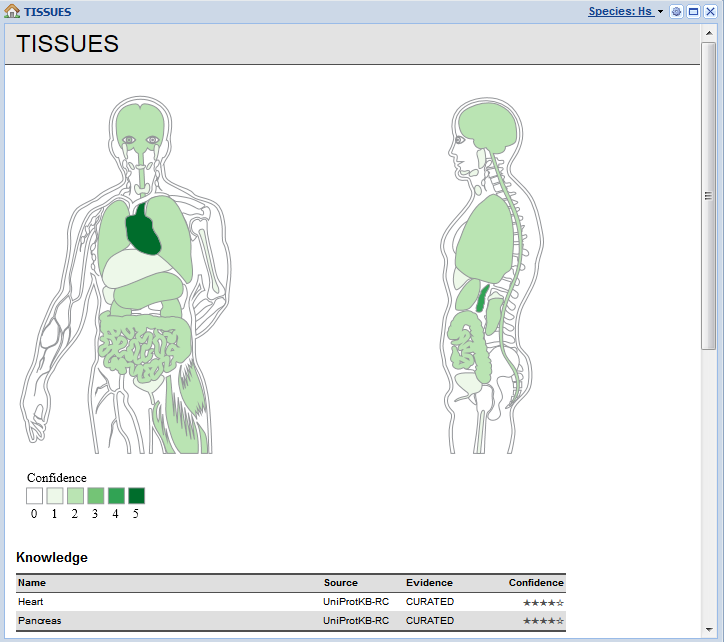
Tissue expression derived from curated knowledge, transcriptomics, proteomics, and automatic text mining.
- Why is your tool unique and special?
What makes TISSUES different from other expression resources is that we have gone to great length to integrate and normalize all available data. Genes/proteins are normalized to the same identifiers used in the STRING database and tissues normalized to terms from the Brenda Tissue Ontology. Moreover, all evidences has quality scores associated with them, which too have been normalized to make it easy to compare the evidence despite its heterogeneity.
- Who is your target audience?
The web interface is currently focused at biologists who want to be able to easily look up a protein of interest and get an overview of where in the body it may be expressed. However, we also very much cater to fellow bioinformaticians by making all data available under the Creative Commons Attribution license.
- Why did you create your tool?
The web resource was developed as a spinoff of our research. To analyze protein interaction networks and cellular signaling processes, it is important to have a good handle on which proteins are present where. To this end we needed to collect and standardize the available data.
- What is your greatest success story so far?
That is too early to say since the tool has only just been published. However, it has been very positively received everywhere we’ve presented it, so we are hopeful that many will be using it.
- What improvements are coming in the future?
We have begun to work on extending the scope of TISSUES to cover not only human but also some of the main mammalian model organisms.
- Who is the team behind your knowledge base?
The data and text mining of the various sources was done by present and former members of my group (Alberto Santos, Kalliopi Tsafou and Sune Pletscher-Frankild). The resource was developed in collaboration with the group of Sean I. O’Donoghue, who developed the interactive data visualization. The on-going work on extending the resource with model organisms is done in collaboration with the group of Jan Gorodkin.
Thanks to Dr. Jensen, for guiding us through their extremely useful and FREE tool. Be sure to check out their plugin in the plugin library.

What is the prominent advantage of this tool?