This is the first blog post in a series on our Gene Wiki renewal. More details below.
First, let’s recap a bit of history on our Gene Wiki project. We originally proposed the Gene Wiki as one aim of our NIH grant to develop BioGPS, an crowdsourced online portal for aggregating gene-centric information. As part of a Master’s student’s thesis project, we created ~7500 Wikipedia “stubs” on human genes. Our Wikipedia bot basically aggregated gene information from many gene databases, reformatted it into a useful infobox template, and brought all gene pages up to a systematic level of content.1
We intended for the Gene Wiki to be a one-and-done project, a cute offshoot that leveraged lots of data integration work we’d already done for BioGPS. But it became clear that the Gene Wiki justified a bigger project of its own. We submitted an R01 proposal to further develop the Gene Wiki, which was eventually funded by NIGMS starting in 2010 for four years.
The first three years of that grant period are now complete, and we are asking the NIH for continued support for another grant period. And the first thing we had to convince reviewers of is that the first round of funding was successful.
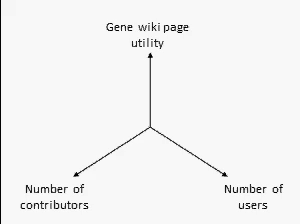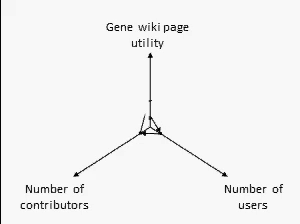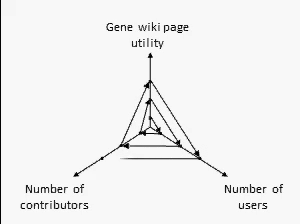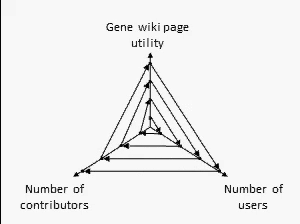 There are many ways to assess the success of the Gene Wiki initiative. But we think the best metric of success by far is examine the positive feedback loop shown at right. In short, your resource must provide a basic level of utility, which then will attract some number of users over time. Some number of users (even a small percentage) will make an edit, ranging from fixing a typo to summarizing a recent paper. That edit increases the utility of the page, which then draws more users and more editors.
There are many ways to assess the success of the Gene Wiki initiative. But we think the best metric of success by far is examine the positive feedback loop shown at right. In short, your resource must provide a basic level of utility, which then will attract some number of users over time. Some number of users (even a small percentage) will make an edit, ranging from fixing a typo to summarizing a recent paper. That edit increases the utility of the page, which then draws more users and more editors.
We think this positive feedback loop defines all successful crowdsourcing projects.2 So, briefly, in order:
- Utility: This axis is the most difficult to quantify, but we believe that the utility of each Gene Wiki page is based on aggregating and integrating information from a diverse set of genomic resources, including NCBI, Ensembl, UniProt, PDB, Gene Ontology, etc.
- Usage: The Gene Wiki in aggregate was viewed 68 million times in the last year. Averaged out over the entire year, that’s over two page views every single second.
- Contributions: Human editors have contributed over 15,000 edits in the last year, and a roughly equivalent number of edits were made by computer bots.
Sure, we also worked on other stuff during the first project period, including automated detection of vandalism3, text mining of structured annotations4, and mashing up the Gene Wiki with other crowdsourced resources5. But ultimately, even if we successfully did all of that but didn’t have a robust critical mass of users and editors, the project would have been a failure.
So we think we have made a strong case that our progress during the first funding period was successful. The following posts will cover what we intend to do during the next funding period.
This blog post is part of a series of entries on our NIH proposal to continue developing the Gene Wiki. The other posts are here:
Post #0: Introduction
Post #1: Gene Wiki progress report (this post)
Post #2: Aim 1: Diseases and drugs
Post #3: Aim 2: Outreach
Post #4: Aim 3: Centralized Model Organism Database
Post #5: Aim 4: Patient-aligned crowdsourcing

Trackbacks/Pingbacks