As we develop BioGPS, many people have asked, why develop another gene portal? With all the already-existing websites that display gene annotation, where does BioGPS fit into this crowded landscape?
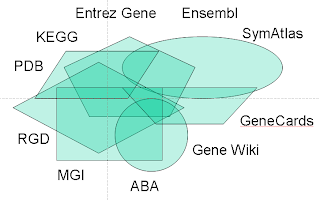 I think this question begs a closer look at why there are so many gene portals in existence to begin with. Suppose you are interested in learning all that is known about the classic cell-cycle gene CDK2. You might start first with NCBI’s Entrez Gene and EBI’s Ensembl. Then you’d want to look at the model organism databases – MGI and RGD. You’d probably want to check an annotation aggregator like GeneCards. And then data providers like SymAtlas and the Allen Brain Atlas. Definitely can’t forget the Gene Wiki either. And on and on, down to tiny and (relatively) unknown resources run out of small academic labs.
I think this question begs a closer look at why there are so many gene portals in existence to begin with. Suppose you are interested in learning all that is known about the classic cell-cycle gene CDK2. You might start first with NCBI’s Entrez Gene and EBI’s Ensembl. Then you’d want to look at the model organism databases – MGI and RGD. You’d probably want to check an annotation aggregator like GeneCards. And then data providers like SymAtlas and the Allen Brain Atlas. Definitely can’t forget the Gene Wiki either. And on and on, down to tiny and (relatively) unknown resources run out of small academic labs.
All of these databases have basically the same goal – to display gene annotation information. And while each displays some unique information that you wouldn’t want to miss, it’s also plainly clear that lots of annotation and functionality is shared between these sites. My estimate? 80% duplication for 20% innovation…
How did we find ourselves in this fragmented landscape of gene portals? I believe in most cases (at least for SymAtlas and expression.gnf.org before that), these resources evolve out of the same basic chronology:
- researchers generate a new annotation source (microarray data set, computational predictions, etc.)
- researchers investigate ways to add these new data to existing resources to share with the world, but find no easy/flexible way to do it
- researchers build simple web site to display new data
- web site evolves to handle classic “gene portal” tasks, including searching, synonym resolution, and annotation display
Wouldn’t it be great if we in the bioinformatics developer community could just focus on that 20% innovation? More on how BioGPS enables this in a future post…

That’s it exactly
Database of Genomic Variants is particularly sparse…..and it always will be if they limit themselves to published manuscripts. Maybe we need something a little less formal where clinicians can upload their observations/findings. We’ve been trying to figure out how to incorporate microarray into clinical practice particularly with regards to the population with mental retardation. The most frequent finding is something novel that has never been reported and may not be related to the phenotype that we’re observing.
It shouldn’t be that difficult to develop something simple and functional in these early days of microarray. Standard texts made use of case-series that simply may not be practical for rare, sporadic or novel variants. The Wikipedians are building gene pages: perhaps we could use the “discussion” tab of the various Wikipedia articles to upload observations. We might upload a consent form to the wikipedian “standards for information on living persons.” It might be a crude start, but what better way to connect patients, clinicians, bioinformatics, and bench researchers?
What do you think?
Paul
aka doctorwolfie on wikipedia
Paul, thanks for your interesting comment. Always good to get the clinical perspective. I think the idea of having a tool for sharing preliminary research findings is interesting and worthwhile. I think Wikipedia, however, is probably not the right forum. (See WP:NOR and WP:V.)
But, I think this would fit in great as a custom BioGPS plugin. Maybe someone could create a custom Mediawiki instance that anyone can contribute to. That might work pretty well. Volunteers?
(Of course, as with all community intelligence initiatives, the hardest part will be convincing the clinicians and researchers to participate.)