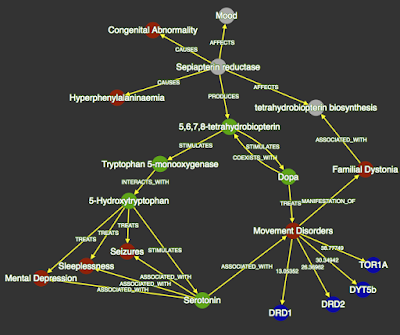
 |
| Personal knowledge graph showing literature-derived connections between Sepiapterin Reductase (SPR) and 5-Hydroxytryptophan (a treatment for patients with deleterious mutations in SPR. |
Benjamin M. Good, Ph.D.1; Richard M. Bruskiewich, Ph.D. 2; Kenneth C. Huellas-Bruskiewicz2; Farzin Ahmed2; Andrew I. Su, Ph.D.1
PubMed now indexes roughly 25 million articles and is growing by more than a million per year. The scale of this “Big Knowledge” repository renders traditional, article-based modes of user interaction unsatisfactory, demanding new interfaces for integrating and summarizing widely distributed knowledge. Natural language processing (NLP) techniques coupled with rich user interfaces can help meet this demand, providing end-users with enhanced views into public knowledge, stimulating their ability to form new hypotheses.
Knowledge.Bio provides a Web interface for exploring the results from text-mining PubMed. It works with subject, predicate, object assertions (triples) extracted from individual abstracts and with predicted statistical associations between pairs of concepts. While agnostic to the NLP technology employed, the current implementation is loaded with triples from the SemRep-generated SemmedDB database and putative gene-disease pairs obtained using Leiden University Medical Center’s ‘Implicitome’ technology.
Users of Knowledge.Bio begin by identifying a concept of interest using text search. Once a concept is identified, associated triples and concept-pairs are displayed in tables. These tables have text-based and semantic filters to help refine the list of triples to relations of interest. The user then selects relations for insertion into a personal knowledge graph implemented using cytoscape.js. The graph is used as a note-taking or ‘mind-mapping’ structure that can be saved offline and then later reloaded into the application. Clicking on edges within a graph or on the ‘evidence’ element of a triple displays the abstracts where that relation was detected, thus allowing the user to judge the veracity of the statement and to read the underlying articles.
Knowledge.Bio is a free, open-source application that can provide, deep, personal, concise, shareable views into the “Big Knowledge” scattered across the biomedical literature. It is available at http://knowledge.bio, with source code at https://bitbucket.org/starinformatics/gbk.

The visual model is far superior for people that think visually. It allows them to use the part of their brain that they use best.
This tool is excellent for visual learners (Interactive tool for literature – based discovery) It allows people that are visual to do what they do best see the best solution from the data
.