When it comes to packing sleeping bags, it seems like there are two kinds of people. Some people meticulously squeeze out every drop of air, rolling the bag into a perfect cylinder. Others impatiently stuff the bag willy-nilly into a sack, and with occasional shoves from a helpful foot (or two), end up with a misshapen lump. This is all well and good for a sleeping bag… but how do you even begin to pack two meters of DNA1 into a single nucleus? And how does a million meters of genetic material compress into the size of a single poppyseed?2
All that makes us human is compacted into 23 pairs of these stubby, insignificant-looking things. [By Abogomazova, CC BY-SA 3.0]
DNA’s double helix may have been one of the greatest discoveries of the last century, but one of today’s most intriguing mysteries is the loopome—the 3-D architecture of the genome. Arranged into patterns of 10,000+ loops, the structure of the genome is anything but haphazard.3 Many of these loops, defined by evolutionarily conserved start and end sites, are preserved in genomes across many species.4
Loops are formed, in part, by CTCF—an architectural protein that binds to DNA and other CTCF proteins. When two CTCF proteins bind in a specific way, they can create a loop by connecting two distant sites within the genome. CTCF binds DNA at specific sites, forming loops that coordinate long-range interactions between DNA regulatory elements.5 This can facilitate transcription if enhancers are brought close to their target promoters, but can also block gene expression by separating regulatory elements in the loopome. Further, CTCF can define boundaries of topologically associating domains (TADs) in the genome.6 These TADs are like neighborhoods of clustered DNA that don’t really interact with DNA outside of their own TAD.7 Moreover, these regions are comprised of genes that are typically co-regulated, since TADs generally share similar chromatin modifications.8
With 55,000-65,000 binding sites present across mammalian genomes,9 CTCF’s effect on gene expression is sure to impact all areas of cellular function. While complete depletion of CTCF in mice is fatal,10 human mutations in CTCF have been associated with anything from cancers11 to brain abnormalities.12 And while investigation of CTCF could lead anywhere, one of the most fascinating possibilities is its potential role in generating cell type diversity. All cells contain the same DNA, but can end up as heart cells or neurons—depending on which genes are expressed. Each cell type has a strictly regulated transcriptional program, which may be reflected (or instructed) by the genome structure… especially since there’s increasing evidence for cell-type-specific loopomes.13
Somewhere in that apparent tangle of DNA are distinct patterns of organization, and indeed, the consistent, deliberate architecture of the genome could just be the answer to how DNA instructs all the diversity of life. How to pack a sleeping bag might reveal one’s personality, but how to stuff a nucleus could very well define us all.
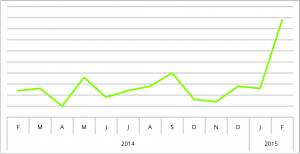
Since December, when Rao and colleagues published their seminal work on the 3D structure of the genome, interest in CTCF has skyrocketed. With this trend, the NIH definitely agrees—they’ve launched a $120 million initiative, called the 4D Nucleome, to study genome architecture across normal development, and in disease.
And if I haven’t convinced you how important genome structure (and CTCF) is, this perfect, cute, and transcendent example of effective scicomm says it all:
REFERENCES:
- Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van IJcken WF, Grosveld FG, Ren B,Wendt KS. (2014) Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A 111(3):996-1001. [↩]
- Amato, Ivan. “Genetic Geometry Takes Shape” QUANTA Magazine (25 Feb 2015). Retrieved 13 Mar 2015 from https://www.quantamagazine.org/20150225-genetic-geometry-takes-shape/ [↩]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159(7):1665-80. [↩]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159(7):1665-80. [↩]
- Merkenschlager M, Odom DT. (2013) CTCF and cohesin: linking gene regulatory elements with their targets. Cell 152(6):1285-97. [↩]
- Ong CT, Corces VG. (2014) CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 15(4):234-46. [↩]
- Ong CT, Corces VG. (2014) CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 15(4):234-46. [↩]
- Bonora G, Plath K, Denholtz M. (2014) A mechanistic link between gene regulation and genome architecture in mammalian development. Curr Opin Genet Dev 27:92-101. [↩]
- Ong CT, Corces VG. (2014) CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 15(4):234-46. [↩]
- Gregor A, Oti M, Kouwenhoven EN, Hoyer J, Sticht H, Ekici AB, Kjaergaard S, Rauch A, Stunnenberg HG, Uebe S, Vasileiou G, Reis A, Zhou H, Zweier C. (2013) De novo mutations in the genome organizer CTCF cause intellectual disability. Am J Hum Genet 93(1):124-31. [↩]
- Marshall AD, Bailey CG, Rasko JE. (2014) CTCF and BORIS in genome regulation and cancer. Curr Opin Genet Dev 24:8-15. [↩]
- Gregor A, Oti M, Kouwenhoven EN, Hoyer J, Sticht H, Ekici AB, Kjaergaard S, Rauch A, Stunnenberg HG, Uebe S, Vasileiou G, Reis A, Zhou H, Zweier C. (2013) De novo mutations in the genome organizer CTCF cause intellectual disability. Am J Hum Genet 93(1):124-31. [↩]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159(7):1665-80. [↩]
