Be Thankful for the Little Things… and the big things that make the little things work
I’ve said it before: small things can make a big difference. This Thanksgiving (and every day), it’s important that we remember and give thanks for the little things. The little things, like microRNAs (miRNAs).
miRNAs are small, non-coding RNAs that are important for the inhibition of gene expression via RNA interference (RNAi). For RNAi to occur, miRNAs or other small RNA molecules, like small interfering RNAs (siRNAs), must bind to target messenger RNA (mRNA). Once bound, they prevent translation by cleaving or blocking access to the target mRNA. For details, see the pretty animation below!1
But where do these miRNAs and siRNAs come from? Well, if you jump to 2:45 in the video, you’ll meet the special protein, Dicer, encoded by our Gene-of-the-Week: DICER1. Dicer doesn’t actually produce the small RNAs from scratch, but it is necessary for their processing from double-stranded RNA (viral) and pre-miRNAs (encoded by the genome). It’s kind of like how my mom is the one who makes the pumpkin pie, but I, too, deserve credit for slicing it… right? Anyway, Dicer is also important for loading these mature miRNAs onto argonaute proteins, necessary for the formation of the RNA-induced silencing complex, or RISC.2
Below is a slightly intimidating figure from Foulkes, et al. (2014) that shows the structure of Dicer (b and c), as well as several common mutations within DICER1 (a).
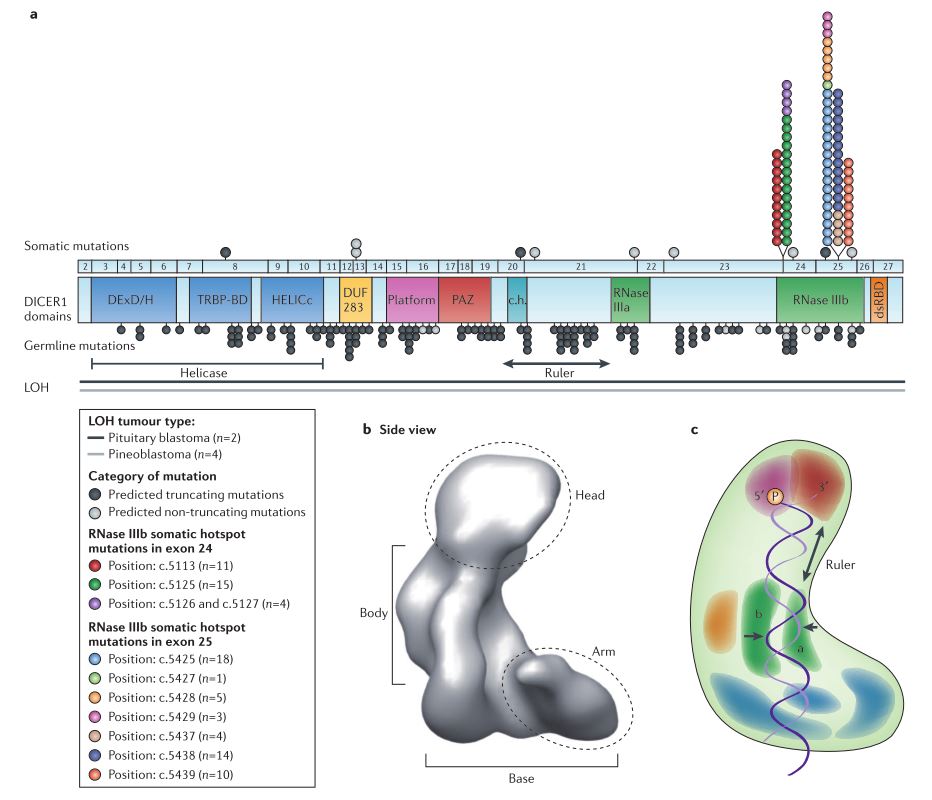
(a) 133 reported germline and 95 proven somatic DICER1 mutations. Several important domains are color-coded along the DICER1 strand: the DExD/H box helicase domain locates and clamps down on dsRNA; Platform and PAZ form binding pockets for the 3′-overhang and 5′-phosphate ends of the dsRNA; the connector helix (c.h.) is part of the linker/ruler that determines the size of the small RNA produced by the cleavage; RNase IIIa and RNase IIIb form the catalytic core responsible for cleaving the dsRNA. (b) and (c) show the structure of Dicer with the important domains color-coded as in (a). From Foulkes et al. (2014)
Mutations in DICER1 can cause what is called “DICER1 syndrome,” which is inherited in an autosomal dominant pattern. Despite the many dots in (a) of the above figure, DICER1 syndrome is thankfully quite rare. The number of associated cancers, however, is huge. DICER1 mutations have been associated with lung, breast, skin, ovarian, and oral cancers. The most common phenotype associated with DICER1 syndrome is pleuropulmonary blastoma (of the lungs). Additional phenotypes include Wilms’ tumor, Sertoli-Leydig cell tumors, and cystic nephroma, all of which are also thankfully very rare and treatable. So far, it’s unknown how the mutations and phenotypes are linked. In fact, both decreased and increased levels of DICER1 have been associated with cancers, so it is unclear if it can be classified oncogene or tumor suppressor gene!3
Recently, Dias et al. (2014) reported that Dicer may play an important role in the “pro-resilient” effect of β-catenin.4 Briefly, increased β-catenin (Ctnnb1) expression in the nucleus accumbens of mice decreases their depressive responses in social defeat and forced-swim tests. β-catenin overexpression also produces anxiolytic effects in the elevated plus maze, while leaving preference for sucrose and cocaine unaffected. Dias et al. reported low β-catenin levels in the post-mortem nucleus accumbens of depressed humans. They also found that β-catenin binds to Dicer1, modulating miRNA production. They found that mice without Dicer1 did not benefit from β-catenin overexpression. Therefore, the pro-resilient effect of β-catenin may be mediated through Dicer. The Dicer connection between resilience, β-catenin, and miRNAs may provide targets for depression treatment in the future!
So this Thanksgiving, be thankful for Dicer, which is important for making important small RNAs that might make us more resilient against the things that we should not think about on Thanksgiving…
References:
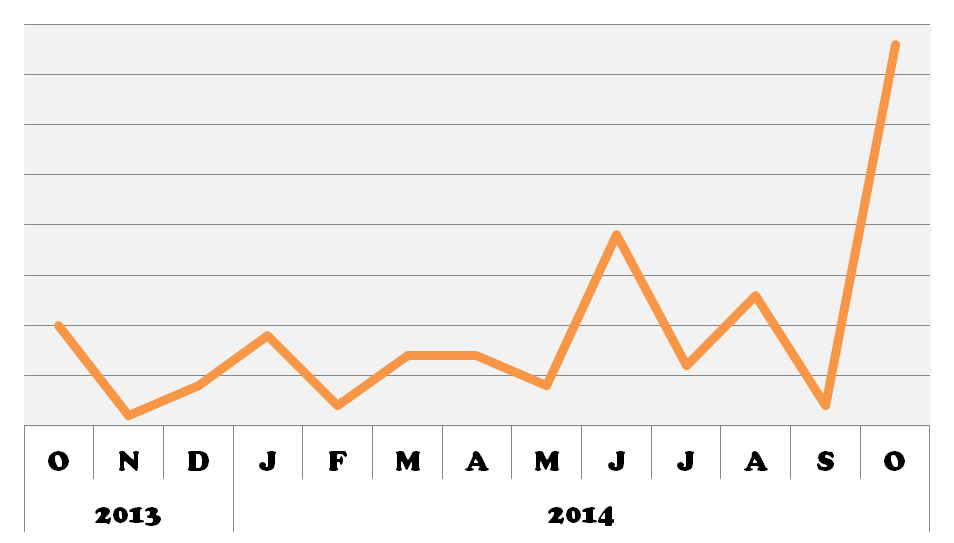
DICER1 popularity increased in October, potentially spurred on by the comprehensive review by Foulkes et al. published in September. It will be interesting to see how its popularity has increased this month as a result of the stress resiliency findings from Dias et al.!
- Fire, A., Xu, S., Montgomery, M., and Kostas, S. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [↩]
- Foulkes, W.D., Priest, J.R., and Duchaine, T.F. (2014). DICER1: mutations, microRNAs and mechanisms. Nat. Rev. Cancer 1–11. [↩]
- Vogelstein, B., Papadopoulos, N., Velculescu, V.E., Zhou, S., Diaz, L. a, and Kinzler, K.W. (2013). Cancer genome landscapes. Science 339, 1546–1558. [↩]
- Dias, C., Feng, J., Sun, H., Shao, N.Y., Mazei-Robison, M.S., Damez-Werno, D., Scobie, K., Bagot, R., LaBonté, B., Ribeiro, E., et al. (2014). β-catenin mediates stress resilience through Dicer1/microRNA regulation. Nature. [↩]

