We are happy to release a new version of BioGPS with some exciting new features:
1. Unified query interface
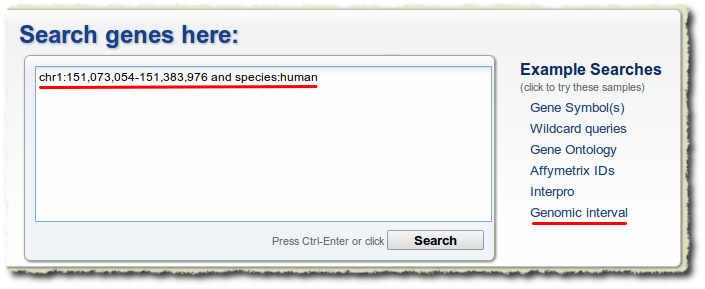
BioGPS previously had two search query interfaces. The “simple” query interface (the default shown on the home page) handled the vast majority of user searches and restricted the search to gene symbols or identifiers. We also offered an “Advanced search” query interface for “keyword” searches (matching both identifiers and text descriptions like gene names or gene summaries) or genome interval search.
We have now made the “simple” query interface into a unified omnibox for gene queries. Any search terms you enter will automatically be searched against the entire gene record, and matches to the gene symbol or gene identifiers will simply be weighted higher in the search results. In addition, genome interval queries are available too using a syntax like this: “chr1:151,073,054-151,383,976 and species:human“.
2. User-customized species preferences
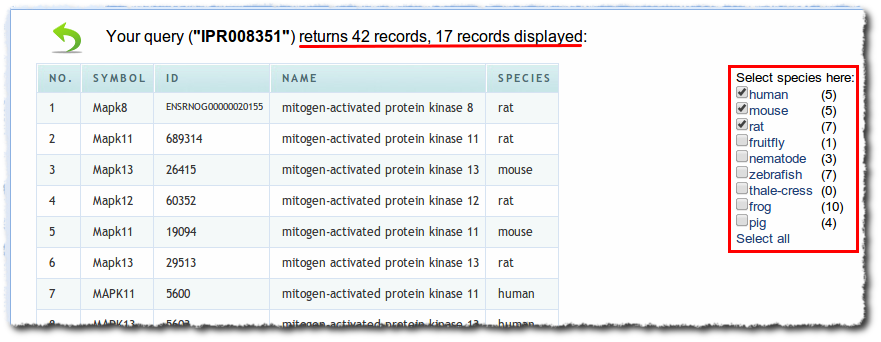
BioGPS’s roots are in human, mouse, and rat genes, but over time and by user request, we’ve added support for many other species. Currently supported species also include fruitfly, nematode, zebrafish, frog and pig (more details here). Although the increased coverage is good for our user base as a whole, each individual user is only interested in the specific list of species that is relevant to them. Therefore, we added a new feature to allow users to limit search results only from the species they picked. When users toggle their species selection, the changes will be saved automatically, so that it will be remembered for future queries.
Besides two visible changes above, the underlying gene query engine has been upgraded significantly as well, which leads to more accurate query results. And the infrastructure is now much more scalable so that we can easily expand our list of supported species. If you have your favorite species you would like to be included in BioGPS, please give your vote by emailing us.
Enjoy!