This guest post was written by Adriel Carolino, a summer intern who has been spearheading this project to create a Centralized Model Organism Database (CMOD).
The recent explosion of metagenomic sequencing has resulted in an immense expanse of microbial genetic information. Current metagenomic analyses typically revolve around a taxonomic survey — describing what microbial species are present in a given sample. However, the field is gradually moving toward more functional analyses of the specific genes and gene products expressed by those microbes.
Unfortunately, there currently isn’t a single public resource that systematically catalogues all microbial gene annotations. For example, there is no database (that we are aware of) that you can query for “all species in the Firmicutes phylum that have a gene involved in potassium ion transport and in a nutrient sensing pathway”. There are two challenges with creating such a system.
First, we need to create a database that can handle the large scale and heterogeneity of microbial gene annotation data. Recently, we have begun exploring Freebase as a platform on which to build such a system. Freebase is a graph database with a structure characterized by sets of interlinked related nodes. Freebase allows for the storage of heterogeneous data in a structured manner; a node can have limitless connectivity while still upholding the intricacy of each connection. A Freebase-backed microbial database would allow us to amass and integrate information from a variety of different annotation projects.
Second, we need to populate that database with annotations from many different sources. Freebase also excels in this regard because it enables the Long Tail of scientists to contribute to the metagenomic annotation process. Like Wikipedia, Freebase welcomes contributions from the community, anything from individual facts from stand-alone labs to large data imports from large-scale efforts. The many varying groups within the community would all be able to input their information, and these contributions would have the ability to be queried for both as a whole and separately. The different microbes could be compared and grouped according to a variety of properties with modifiable specificity.
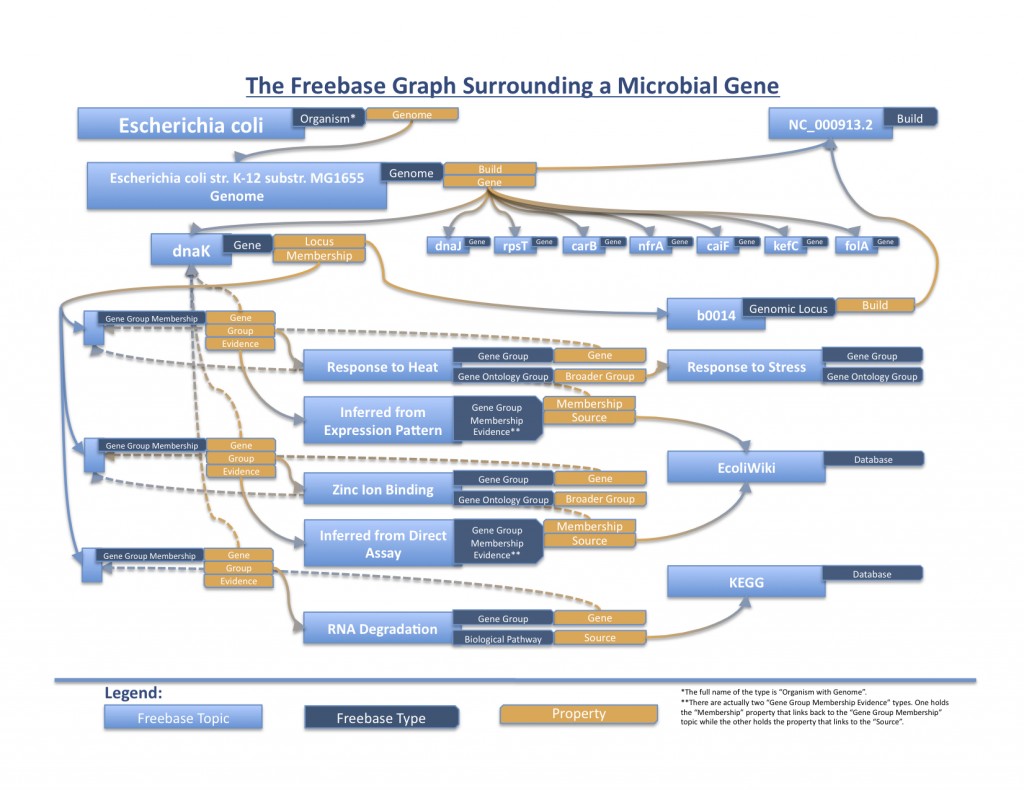
We first constructed a comprehensive data model that has been filled in with info from the well-studied organisms Escherichia coli K-12 substr. MG1655 and Pseudomonas aeruginosa PAO1. With information obtained from NCBI, the Gene Ontology, the UCSC Microbial Genome Browser, EcoliWiki, EcoCyc, and the Pseudomonas Genome Database, we have created 70 gene topics (35 from each genome) so far and are in the process of inputting more. They can all be viewed at the Microbial Gene Base within Freebase.
Right now, this basic infrastructure enables simple queries to, for example, retrieve all E. coli K-12 MG1655 genes that are involved in “zinc ion binding”. The Gene Ontology term is linked to a group of genes via sourced evidence, and each individual gene can point to properties it holds such as its genome, NCBI Entrez Gene ID, and locus (along with its specific location in a build).
We have obtained data from KEGG and EcoCyc regarding biological pathways and will upload that information soon. We hope to include other relevant data and other microbial species into the Freebase graph as well. We also believe, success pending, that an awesome and useful future aspect would be the development of a generic cross-species genome browser that could use and display the information for any species in the Microbial Gene Base.
Why do we think this system is important, especially since there is already a Generic Model Organism Database (GMOD) project? Because we think it’s unlikely that every microbial species will have a large enough community to justify its own GMOD instance (especially given the developer and curator requirements). For the Long Tail of microbial species, we think this Centralized Model Organism Database (CMOD) will be an effective, efficient, and scalable method to collaboratively organize knowledge on microbial genes.