As the end of 2011 was a busy time grant-writing, I didn’t get an opportunity to highlight several recent publications that came out from our lab. Better late than never:
- Hu Y, Galkin AV, Wu C, Reddy V, Su AI (2011) CAFET Algorithm Reveals Wnt/PCP Signature in Lung Squamous Cell Carcinoma.PLoS ONE 6(10): e25807. (Full text)
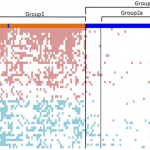 This paper described CAFET, a novel algorithm for enrichment-based analyses of genome-scale data. Traditional enrichment algorithms identify enrichment of functional groups among differentially-expressed genes. In contrast, CAFET works on the ortholonal axis by seeking enrichment of clinically-relevant sample groups among all samples bearing particular gene expression signatures. We applied CAFET to two large cohorts of lung cancer patients to implicate a non-canonical branch of the Wnt pathway in squamous cell carcinoma.
This paper described CAFET, a novel algorithm for enrichment-based analyses of genome-scale data. Traditional enrichment algorithms identify enrichment of functional groups among differentially-expressed genes. In contrast, CAFET works on the ortholonal axis by seeking enrichment of clinically-relevant sample groups among all samples bearing particular gene expression signatures. We applied CAFET to two large cohorts of lung cancer patients to implicate a non-canonical branch of the Wnt pathway in squamous cell carcinoma.
- Good BM, Clarke EL, de Alfaro L, Su AI (2012) The Gene Wiki in 2011: community intelligence applied to human gene annotation. Nucleic Acids Res., 40(1):D1255-61. (Full text)
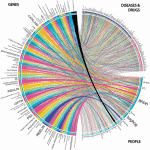 This paper described provided an update on the overall status of the Gene Wiki project. In addition to improvements to the underlying infrastructure, we also recapped the content statistics (1.42 million words in 10369 articles), usage rates (50 million pageviews per year), and editing statistics (17,000 per year). Moreover, we introduce the use of the WikiTrust to better evaluate the quality of community-contributed gene annotation data. Note that the image at left was selected for the cover of the Database Issue (high-res: 929 KB, vector: 6 MB).
This paper described provided an update on the overall status of the Gene Wiki project. In addition to improvements to the underlying infrastructure, we also recapped the content statistics (1.42 million words in 10369 articles), usage rates (50 million pageviews per year), and editing statistics (17,000 per year). Moreover, we introduce the use of the WikiTrust to better evaluate the quality of community-contributed gene annotation data. Note that the image at left was selected for the cover of the Database Issue (high-res: 929 KB, vector: 6 MB).
- Good BM, Howe DG, Lin SM, Kibbe WA, Su AI (2011) Mining the Gene Wiki for functional genomic knowledge. BMC Genomics, 12(1):603. (Full text)
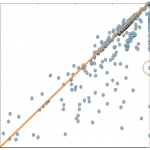 The Gene Wiki has clearly demonstrated a critical mass of editors who contribute unstructured knowledge about gene function. This paper sought to mine those unstructured contributions for structured data (which are then amenable to computation and statistics). We used basic text mining to identify candidate novel annotations, then partnered with professional biocurators to evaluate the accuracy of this approach.
The Gene Wiki has clearly demonstrated a critical mass of editors who contribute unstructured knowledge about gene function. This paper sought to mine those unstructured contributions for structured data (which are then amenable to computation and statistics). We used basic text mining to identify candidate novel annotations, then partnered with professional biocurators to evaluate the accuracy of this approach.
- Good BM, and Su AI (2011) Games with a scientific purpose. Genome Biol., 12(12):135. (Full text)
 This commentary described several recent scientific advances based on crowdsourcing through games. The Foldit team used this novel and emerging approach to predict novel protein structures and to design new protein folding algorithms. Our group is also interested in using games to advance genetics and genomics, including expanding our prototype Dizeez game.
This commentary described several recent scientific advances based on crowdsourcing through games. The Foldit team used this novel and emerging approach to predict novel protein structures and to design new protein folding algorithms. Our group is also interested in using games to advance genetics and genomics, including expanding our prototype Dizeez game.
All papers are freely accessible using the full text links above.
